Authors:
Mannochio-Russo, Helena [1, 2] ; Bueno, Paula Carolina P. [3, 4] ; Bauermeister, Anelize [1, 5] ; de Almeida, Rafael Felipe [6] ; Dorrestein, Pieter C. [1] ; Cavalheiro, Alberto Jose [2] ; Bolzani, Vanderlan S. [2]
Abstract:
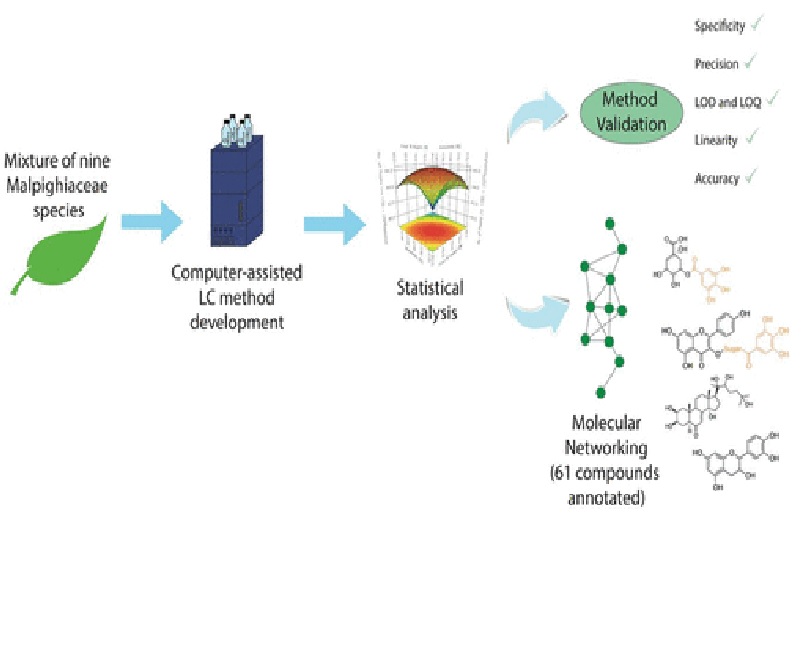
Proper chromatographic methods may reduce the challenges inherent in analyzing natural product extracts, especially when utilizing hyphenated detection techniques involving mass spectrometry. As there are many variations one can introduce during chromatographic method development, this can become a daunting and time-consuming task. To reduce the number of runs and time needed, the use of instrumental automatization and commercial software to apply Quality by Design and statistical analysis automatically can be a valuable approach to investigate complex matrices. To evaluate this strategy in the natural products workflow, a mixture of nine species from the family Malpighiaceae was investigated. By this approach, the entire data collection and method development procedure (comprising screening, optimization, and robustness simulation) was accomplished in only 4 days, resulting in very low limits of detection and quantification. The analysis of the individual extracts also proved the efficiency of the use of a mixture of extracts for this workflow. Molecular networking and library searches were used to annotate a total of 61 compounds, including O-glycosylated flavonoids, C-glycosylated flavonoids, quinic/shikimic acid derivatives, sterols, and other phenols, which were efficiently separated by the method developed. These results support the potential of statistical tools for chromatographic method optimization as an efficient approach to reduce time and maximize resources, such as solvents, to get proper chromatographic conditions.
1 Pharmaceutical Sciences, University of California, San Diego, La Jolla, California 92093, United States
2 NuBBE, Department of Biochemistry and Organic Chemistry, Institute of Chemistry, São Paulo State University (UNESP), 14800-901, Araraquara, SP Brazil
3 Max Planck Institute of Molecular Plant Physiology, 14476, Potsdam-Golm, Germany
4 Faculty of Pharmaceutical Sciences of Ribeirão Preto, Department of Physics and Chemistry, University of São Paulo, 14049-900, Ribeirão Preto, SP Brazil
5 Biomedical Sciences Institute, University of São Paulo, 05508-900 São Paulo, SP Brazil
6 Department of Biological Sciences, Lamol Lab, Feira de Santana State University (UEFS), Feira de Santana, BA 44036-900, Brazil
Link to article: https://pubs.acs.org/doi/10.1021/acs.jnatprod.0c00495